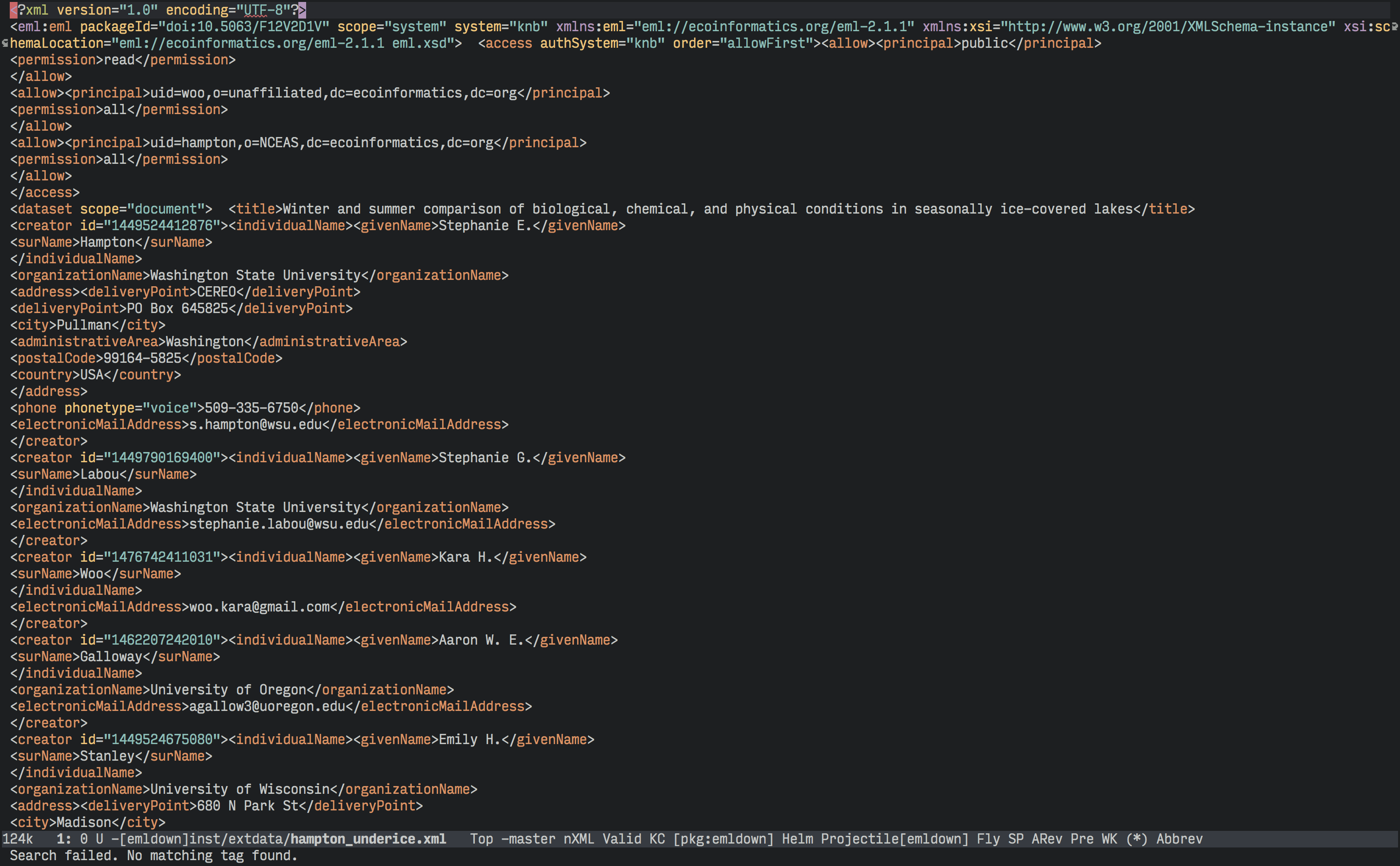
There is no problem in science quite as frustrating as other peoples' data. Whether it's malformed spreadsheets, disorganized documents, proprietary file formats, data without metadata, or any other data scenario created by someone else, scientists have taken to Twitter to complain about it. As a political scientist who regularly encounters so-called "open data" in PDFs, this problem is particularly irritating. PDFs may have "portable" in their name, making them display consistently on various platforms, but that portability means any information contained in a PDF is irritatingly difficult to extract computationally. Encountering "open data" PDFs therefore makes me shout things like this repeatedly:
HEY US GOVERNMENT! Tables in PDF documents aren't "Open Data." Please provide machine-readable formats or it doesn't count.
— Anthony A. Boyles (@AABoyles) September 15, 2016
What can we do about such data other than extract it by hand? One answer is rely on tabulizer a package I submitted to rOpenSci that reduces some and often all of the hassle of extracting tabular data locked inside PDFs.
What is tabulizer?
tabulizer provides R bindings to the tabula-java library, the open-source java library that powers Tabula (source code on GitHub). What this means is that tabulizer relies directly on the underlying java classes that power Tabula without the need for system calls or the need to explicitly install Tabula on your system. (A potential downside is the need to handle intricacies of rJava - R's interface to Java, which I discuss in depth below.)
Tabula is an extremely powerful tool for extracting tabular data locked in PDFs. It's an incredibly valuable tool because the PDF file specification does not have a "table" representation. Instead, PDFs simply represent tables through the fixed positioning of text into rows and columns. Thus, unlike HTML, Word (.docx), or Open Document (.odt) file formats, there is no easy programmatic way to identify a table in a PDF. Tabula thus implements novel algorithms for identifying rows and columns of data and extracting them. tabulizer just provides a thin R layer on top of this power Java code.
Unfortunately, this means that tabulizer is not a universal solution to data trapped in PDFs. In particular, it can only identify and extract tables that are represented as text in a PDF:
Oh no no no no no! Just received #otherpeoplesdata as a 276 page set of printed tables scanned in to a PDF
— Dr Elizabeth Sargent (@esargent184) September 11, 2014
If a PDF is a scan of a document or the table is actually an image embedded in the PDF, tabula - and thus tabulizer - are useless. In those cases, users might want to check out the OCR functionality of tesseract, which Jeroen Ooms developed for rOpenSci and discussed previously on this blog.
But it does mean that a substantial amount of difficult-to-parse tabular information in PDFs is now readily and quickly accessible via just one tabulizer function: extract_tables().
Installation
tabulizer is not yet on CRAN. (It's CRAN-ready but due to some underlying developments that are ongoing in the tabula-java library, I'm waiting for a formal release.) In the meantime, it's possible to install tabulizer directly from GitHub.
Before doing that, I would encourage users to make sure they have rJava installed (from CRAN) and that it works correctly on their platform. A lot of users report difficulties installing tabulizer that ultimately boil down to being Java and rJava issues that need to be resolved first. The package README provides a number of details on installation, which requires a strictly ordered set of steps:
- Install the Java Development Kit, if you don't already have it on your system. (Note that the JDK is different from the Java Runtime Environment (JRE) that you almost certainly already have.) Details of how to do this vary a lot between platforms, so see the README for details.
- Install rJava using
install.packages("rJava")and resolve any issues surrounding theJAVA_HOMEenvironment variable that may need to be set before and/or after installing rJava. Again, see the README or various question/answer pairs on StackOverflow for platform-specific instructions. - Install
tabulizerandtabulizerjars(the package containing the tabula java library) using your favorite GitHub package installer:
library("ghit")
ghit::install_github(c("ropensci/tabulizerjars","ropensci/tabulizer"))This should work. If not, set verbose = TRUE in ghit::install_github() to identify the source of any issues. Some common problems are the dependency on the png package, which might need to be installed first. On Windows (depending on your version of R and how it was installed) may require setting INSTALL_opts = "--no-multiarch" in ghit::install_github().
If none of these steps work, scroll through the GitHub issues page for anyone experiencing a similar problem and, if not resolved in any of those discussions, feel free to open an issue on GitHub describing your problem including the fully verbose output of install_github() and your sessionInfo().
Unlocking elections data with tabulizer
Elections data are the bread and butter of a lot of quantitative political science research. Many researchers in my field need to know how many citizens voted and for whom in order to make sense of campaigns, election integrity, partisanship, and so forth. Yet a substantial amount of election-related data is locked in government-produced PDFs. Worse, national, state, and local governments have little to no standardization in the formatting of elections data, meaning even if one could figure out a computational strategy for extracting one kind of data about elections in one year from one state, that computational strategy would likely be useless in the same state in another year or in any other state. Elections provide a fantastic and highly visible example of "open" government data that's not really open or usable at all.
As a simple example, this PDF from the California Secretary of State's office contains historical voter registration and turnout data in a well-formatted table. Why this is a PDF nobody knows. But extracting the tables using tabulizer's extract_tables() function is a breeze with no need to even download the file:
library("tabulizer")
sos_url <-"http://elections.cdn.sos.ca.gov/sov/2016-general/sov/04-historical-voter-reg-participation.pdf"
tab1 <- extract_tables(sos_url)
str(tab1)## List of 2
## $ : chr [1:58, 1:9] "" "Election Date" "Nov. 8, 1910" "Nov. 5, 1912 P" ...
## $ : chr [1:6, 1:9] "" "Election Date" "Nov. 2, 2010" "Nov. 6, 2012 P" ...
The (default) result is a list of two matrices, each containing the tables from pages 1 and 2 of the document, respectively. A couple of quick cleanups and this becomes a well-formatted data frame:
# save header
h <- tab1[[1]][2,]# remove headers in first table
tab1[[1]]<- tab1[[1]][-c(1,2),]# remove duplicated header in second table
tab1[[2]]<- tab1[[2]][-c(1,2),]# merge into one table
tab1df <- setNames(as.data.frame(do.call("rbind", tab1), stringsAsFactors =FALSE), h)
str(tab1df)## 'data.frame': 60 obs. of 9 variables:
## $ Election Date: chr "Nov. 8, 1910" "Nov. 5, 1912 P" "Nov. 3, 1914" "Nov. 7, 1916 P" ...
## $ Eligible : chr "725,000" "1,569,000" "1,726,000" "1,806,000" ...
## $ Democratic : chr "*" "*" "*" "*" ...
## $ Republican : chr "*" "*" "*" "*" ...
## $ Other : chr "*" "*" "*" "*" ...
## $ Total : chr "*" "987,368" "1,219,345" "1,314,446" ...
## $ Total Votes : chr "393,893" "707,776" "961,868" "1,045,858" ...
## $ Registered : chr "*" "71.68%" "78.88%" "79.57%" ...
## $ Eligible : chr "54.33%" "45.11%" "55.73%" "57.91%" ...
Which is very easy to then quickly turn into a time-series visualization of registration rates:
library("ggplot2")
years <-regexpr("[[:digit:]]{4}",tab1df[["Election Date"]])
tab1df$Year <-as.numeric(regmatches(tab1df[["Election Date"]], years))
tab1df$RegPerc <-as.numeric(gsub("%","", tab1df$Registered))## Warning: NAs introduced by coercion
ggplot(tab1df, aes(x = Year, y = RegPerc))+
geom_line()+ ylim(c(0,100))+ ylab("% Registered")+
ggtitle("California Voter Registration, by Year")## Warning: Removed 1 rows containing missing values (geom_path).

Optional arguments
The extract_tables() has several arguments that control extraction and the return value of the function. They performed reasonably well here, but it's worth seeing a few of the other options. The method argument controls the return value. For extremely well-formatted tables, setting this to "data.frame" can be convenient, though it doesn't work perfectly here:
str(tab2 <- extract_tables(sos_url, method ="data.frame"))## List of 2
## $ :'data.frame': 57 obs. of 9 variables:
## ..$ X : chr [1:57] "Election Date" "Nov. 8, 1910" "Nov. 5, 1912 P" "Nov. 3, 1914" ...
## ..$ X.1 : chr [1:57] "Eligible" "725,000" "1,569,000" "1,726,000" ...
## ..$ X.2 : chr [1:57] "Democratic" "*" "*" "*" ...
## ..$ X.3 : chr [1:57] "Republican" "*" "*" "*" ...
## ..$ X.4 : chr [1:57] "Other" "*" "*" "*" ...
## ..$ X.5 : chr [1:57] "Total" "*" "987,368" "1,219,345" ...
## ..$ X.6 : chr [1:57] "Total Votes" "393,893" "707,776" "961,868" ...
## ..$ Turnout : chr [1:57] "Registered" "*" "71.68%" "78.88%" ...
## ..$ Turnout.1: chr [1:57] "Eligible" "54.33%" "45.11%" "55.73%" ...
## $ :'data.frame': 5 obs. of 9 variables:
## ..$ X : chr [1:5] "Election Date" "Nov. 2, 2010" "Nov. 6, 2012 P" "Nov. 4, 2014" ...
## ..$ X.1 : chr [1:5] "Eligible" "23,551,699" "23,802,577" "24,288,145" ...
## ..$ X.2 : chr [1:5] "Democratic" "7,620,240" "7,966,422" "7,708,683" ...
## ..$ X.3 : chr [1:5] "Republican" "5,361,875" "5,356,608" "5,005,422" ...
## ..$ X.4 : chr [1:5] "Other" "4,303,768" "4,922,940" "5,089,718" ...
## ..$ X.5 : chr [1:5] "Total" "17,285,883" "18,245,970" "17,803,823" ...
## ..$ X.6 : chr [1:5] "Total Votes" "10,300,392" "13,202,158" "7,513,972" ...
## ..$ Turnout : chr [1:5] "Registered" "59.59%" "72.36%" "42.20%" ...
## ..$ Turnout.1: chr [1:5] "Eligible" "43.74%" "55.47%" "30.94%" ...
Setting method = "character" returns a list of character vectors with white space reflecting the positioning of text within the PDF's tabular representation:
str(tab3 <- extract_tables(sos_url, method ="character"))## List of 2
## $ : chr "\t\t\t\t\t\t\tTurnout\tTurnout\nElection Date\tEligible\tDemocratic\tRepublican\tOther\tTotal\tTotal Votes\tRegistered\tEligibl"| __truncated__
## $ : chr "\t\t\t\t\t\t\tTurnout\tTurnout\nElection Date\tEligible\tDemocratic\tRepublican\tOther\tTotal\tTotal Votes\tRegistered\tEligibl"| __truncated__
This argument can also be set to "csv", "tsv", or "json" to use a java-level utility to write the table to files in the working directory but this tends to be inconvenient. (For advanced users, method = "asis" returns an rJava object reference for those who want to manipulate the Java representation of the table directly.)
The other most important option to be aware of is guess, which indicates whether a column-finding algorithm should be used to identify column breaks. This should almost always be TRUE, setting it to FALSE will tend to return a less useful structure:
head(extract_tables(sos_url, guess =FALSE)[[1]],10)## [,1]
## [1,] ""
## [2,] ""
## [3,] ""
## [4,] ""
## [5,] "Election Date"
## [6,] "Nov. 8, 1910"
## [7,] "Nov. 5, 1912 P"
## [8,] "Nov. 3, 1914"
## [9,] "Nov. 7, 1916 P"
## [10,] "Nov. 5, 1918"
## [,2]
## [1,] "HISTORICAL VOTER REGISTRATION AND"
## [2,] "PARTICIPATION IN STATEWIDE GENERAL ELECTIONS 1910-2016"
## [3,] "Registration Votes Cast"
## [4,] "Turnout"
## [5,] "Eligible Democratic Republican Other Total Total Votes Registered"
## [6,] "725,000 * * * *393,893*"
## [7,] "1,569,000 * * * 987,368 707,776 71.68%"
## [8,] "1,726,000 * * * 1,219,345 961,868 78.88%"
## [9,] "1,806,000 * * * 1,314,446 1,045,858 79.57%"
## [10,] "1,918,000 * * * 1,203,898 714,525 59.35%"
## [,3]
## [1,] ""
## [2,] ""
## [3,] ""
## [4,] "Turnout"
## [5,] "Eligible"
## [6,] "54.33%"
## [7,] "45.11%"
## [8,] "55.73%"
## [9,] "57.91%"
## [10,] "37.25%"
However, it can be useful if users want to specify the locations of tables manually. The area argument allows users to specifying a c(top,left,bottom,right) vector of coordinates for the location of tables on a page (which is useful if pages also contain other non-tabular content); setting columns with guess = FALSE indicates where the column breaks are within a table. With a little care in specifying column positions we can successfully separate the "P" flags specifying Presidential elections that were earlier concatenated with the election dates:
cols <-list(c(76,123,126,203,249,297,342,392,453,498,548))
tab4 <- extract_tables(sos_url, guess =FALSE, columns = cols)# save header
h <- tab4[[1]][5,-1]# clean tables
tab4[[1]]<- tab4[[1]][-c(1:5,62),-1]
tab4[[2]]<- tab4[[2]][-c(1:5,10:17),-1]# merge into one table
tab4df <- setNames(as.data.frame(do.call("rbind", tab4), stringsAsFactors =FALSE), h)
str(tab4df)## 'data.frame': 60 obs. of 10 variables:
## $ Election Date: chr "Nov. 8, 1910" "Nov. 5, 1912" "Nov. 3, 1914" "Nov. 7, 1916" ...
## $ : chr "" "P" "" "P" ...
## $ Eligible : chr "725,000" "1,569,000" "1,726,000" "1,806,000" ...
## $ Democratic : chr "*" "*" "*" "*" ...
## $ Republican : chr "*" "*" "*" "*" ...
## $ Other : chr "*" "*" "*" "*" ...
## $ Total : chr "*" "987,368" "1,219,345" "1,314,446" ...
## $ Total Votes : chr "393,893" "707,776" "961,868" "1,045,858" ...
## $ Registered : chr "*" "71.68%" "78.88%" "79.57%" ...
## $ Eligible : chr "54.33%" "45.11%" "55.73%" "57.91%" ...
Figuring out columns positions and/or table areas is quite challenging to do by hand, so the locate_areas() provides an interactive interface for identifying areas. It returns lists of coordinates for specific table areas. A higher-level function, extract_areas(), connects that GUI directly to extract_tables() to return the tables within specified areas. Two other functions can be useful in this respect: get_n_pages() indicates the number of pages in a PDF and get_page_dims() indicates the dimensions of the pages.
Some other functionality
In addition to the core functionality around extract_tables(), tabulizer also provides some functions for working with PDFs that might be useful to those trapped in other peoples' data. We'll download the file first just to save some time:
tmp <-tempfile(fileext =".pdf")
download.file(sos_url, destfile = tmp, mode ="wb", quiet =TRUE)The extract_text() function extracts text content of the PDF, separately by page, as character strings:
extract_text(tmp)## [1] "4Election Date Eligible Democratic Republican Other Total Total Votes\r\nTurnout \r\nRegistered\r\nTurnout \r\nEligible\r\nNov. 8, 1910 725,000 * * * * 393,893 * 54.33%\r\nNov. 5, 1912 P 1,569,000 * * * 987,368 707,776 71.68% 45.11%\r\nNov. 3, 1914 1,726,000 * * * 1,219,345 961,868 78.88% 55.73%\r\nNov. 7, 1916 P 1,806,000 * * * 1,314,446 1,045,858 79.57% 57.91%\r\nNov. 5, 1918 1,918,000 * * * 1,203,898 714,525 59.35% 37.25%\r\nNov. 2, 1920 P 2,090,000 * * * 1,374,184 987,632 71.87% 47.26%\r\nNov. 7, 1922 2,420,000 319,107 968,429 244,848 1,532,384 1,000,997 65.32% 41.36%\r\nNov. 4, 1924 P 2,754,000 397,962 1,183,672 240,723 1,822,357 1,336,598 73.34% 48.53%\r\nNov. 2, 1926 2,989,000 410,290 1,298,062 204,510 1,912,862 1,212,452 63.38% 40.56%\r\nNov. 6, 1928 P 3,240,000 592,161 1,535,751 185,904 2,313,816 1,846,077 79.78% 56.98%\r\nNov. 4, 1930 3,463,000 456,096 1,638,575 150,557 2,245,228 1,444,872 64.35% 41.72%\r\nNov. 8, 1932 P 3,573,000 1,161,482 1,565,264 162,267 2,889,013 2,330,132 80.65% 65.22%\r\nNov. 6, 1934 3,674,000 1,555,705 1,430,198 154,211 3,140,114 2,360,916 75.19% 64.26%\r\nNov. 3, 1936 P 3,844,000 1,882,014 1,244,507 127,300 3,253,821 2,712,342 83.36% 70.56%\r\nNov. 8, 1938 4,035,000 2,144,360 1,293,929 173,127 3,611,416 2,695,904 74.65% 66.81%\r\nNov. 5, 1940 P 4,214,000 2,419,628 1,458,373 174,394 4,052,395 3,300,410 81.44% 78.32%\r\nNov. 3, 1942 4,693,000 2,300,206 1,370,069 150,491 3,820,776 2,264,288 59.26% 48.25%\r\nNov. 7, 1944 P 5,427,000 2,418,965 1,548,395 173,971 4,141,331 3,566,734 86.13% 65.72%\r\nNov. 5, 1946 5,800,000 2,541,720 1,637,246 204,997 4,383,963 2,759,641 62.95% 47.58%\r\nNov. 2, 1948 P 6,106,000 2,892,222 1,908,170 261,605 5,061,997 4,076,981 80.54% 66.77%\r\nNov. 7, 1950 6,458,000 3,062,205 1,944,812 237,820 5,244,837 3,845,757 73.32% 59.55%\r\nNov. 4, 1952 P 7,033,000 3,312,668 2,455,713 229,919 5,998,300 5,209,692 86.85% 74.07%\r\nNov. 2, 1954 7,565,000 3,266,831 2,415,249 203,157 5,885,237 4,101,692 69.69% 54.22%\r\nNov. 6, 1956 P 8,208,000 3,575,635 2,646,249 186,937 6,408,821 5,547,621 86.56% 67.59%\r\nNov. 4, 1958 8,909,000 3,875,630 2,676,565 200,226 6,752,421 5,366,053 79.47% 60.23%\r\nNov. 8, 1960 P 9,587,000 4,295,330 2,926,408 242,888 7,464,626 6,592,591 88.32% 68.77%\r\nNov. 6, 1962 10,305,000 4,289,997 3,002,038 239,176 7,531,211 5,929,602 78.73% 57.54%\r\nNov. 3, 1964 P 10,959,000 4,737,886 3,181,272 264,985 8,184,143 7,233,067 88.38% 66.00%\r\nNov. 8, 1966 11,448,000 4,720,597 3,350,990 269,281 8,340,868 6,605,866 79.20% 57.70%\r\nNov. 5, 1968 P 11,813,000 4,682,661 3,462,131 442,881 8,587,673 7,363,711 85.75% 62.34%\r\nNov. 3, 1970 12,182,000 4,781,282 3,469,046 456,019 8,706,347 6,633,400 76.19% 54.45%\r\nNov. 7, 1972 P 13,322,000 5,864,745 3,840,620 760,850 10,466,215 8,595,950 82.13% 64.52%\r\nNov. 6, 1973 S 13,512,000 5,049,959 3,422,291 617,569 9,089,819 4,329,017 47.62% 32.04%\r\nNov. 5, 1974 13,703,000 5,623,831 3,574,624 729,909 9,928,364 6,364,597 64.11% 46.45%\r\nNov. 2, 1976 P 14,196,000 5,725,718 3,468,439 786,331 9,980,488 8,137,202 81.53% 57.32%\r\nNov. 7, 1978 14,781,000 5,729,959 3,465,384 934,643 10,129,986 7,132,210 70.41% 48.25%\r\nNov. 6, 1979 S 15,083,000 5,594,018 3,406,854 1,006,085 10,006,957 3,740,800 37.38% 24.80%\r\nNov. 4, 1980 P 15,384,000 6,043,262 3,942,768 1,375,593 11,361,623 8,775,459 77.24% 57.04%\r\nNov. 2, 1982 15,984,000 6,150,716 4,029,684 1,378,699 11,559,099 8,064,314 69.78% 50.45%\r\nNov. 6, 1984 P 16,582,000 6,804,263 4,769,129 1,500,238 13,073,630 9,796,375 74.93% 59.08%\r\nNov. 4, 1986 17,561,000 6,524,496 4,912,581 1,396,843 12,833,920 7,617,142 59.35% 43.38%\r\nNov. 8, 1988 P 19,052,000 7,052,368 5,406,127 1,546,378 14,004,873 10,194,539 72.81% 53.51%\r\nNov. 6, 1990 19,245,000 6,671,747 5,290,202 1,516,078 13,478,027 7,899,131 58.61% 41.05%\r\nNov. 3, 1992 P 20,864,000 7,410,914 5,593,555 2,097,004 15,101,473 11,374,565 75.32% 54.52%\r\nNov. 2, 1993 S 20,797,000 7,110,142 5,389,313 2,043,168 14,524,623 5,282,443 36.37% 27.73%\r\nNov. 8, 1994 18,946,000 7,219,635 5,472,391 2,031,758 14,723,784 8,900,593 60.45% 46.98%\r\nNov. 5, 1996 P 19,526,991 7,387,504 5,704,536 2,570,035 15,662,075 10,263,490 65.53% 52.56%\r\nNov. 3, 1998 20,806,462 6,989,006 5,314,912 2,665,267 14,969,185 8,621,121 57.59% 41.43%\r\nNov. 7, 2000 P 21,461,275 7,134,601 5,485,492 3,087,214 15,707,307 11,142,843 70.94% 51.92%\r\nNov. 5, 2002 21,466,274 6,825,400 5,388,895 3,089,174 15,303,469 7,738,821 50.57% 36.05%\r\nOct. 7, 2003 S 21,833,141 6,718,111 5,429,256 3,236,059 15,383,526 9,413,494 61.20% 43.12%\r\nNov. 2, 2004 P 22,075,036 7,120,425 5,745,518 3,691,330 16,557,273 12,589,683 76.04% 57.03%\r\nNov. 8, 2005 S 22,487,768 6,785,188 5,524,609 3,581,685 15,891,482 7,968,757 50.14% 35.44%\r\nNov. 7, 2006 22,652,190 6,727,908 5,436,314 3,672,886 15,837,108 8,899,059 56.19% 39.29%\r\nNov. 4, 2008 P 23,208,710 7,683,495 5,428,052 4,192,544 17,304,091 13,743,177 79.42% 59.22%\r\nMay 19, 2009 S 23,385,819 7,642,108 5,325,558 4,185,346 17,153,012 4,871,945 28.40% 20.80%\r\nHISTORICAL VOTER REGISTRATION AND\r\nPARTICIPATION IN STATEWIDE GENERAL ELECTIONS 1910-2016\r\nVotes CastRegistration\r\n5Election Date Eligible Democratic Republican Other Total Total Votes\r\nTurnout \r\nRegistered\r\nTurnout \r\nEligible\r\nNov. 2, 2010 23,551,699 7,620,240 5,361,875 4,303,768 17,285,883 10,300,392 59.59% 43.74%\r\nNov. 6, 2012 P 23,802,577 7,966,422 5,356,608 4,922,940 18,245,970 13,202,158 72.36% 55.47%\r\nNov. 4, 2014 24,288,145 7,708,683 5,005,422 5,089,718 17,803,823 7,513,972 42.20% 30.94%\r\nNov. 8, 2016 P 24,875,293 8,720,417 5,048,398 5,642,956 19,411,771 14,610,509 75.27% 58.74%\r\nNotes\r\n* Indicates information not available. \r\nIn 1911, women gained the right to vote in California.\r\nIn 1972, the voting age was lowered from 21 to 18.\r\nRegistration Votes Cast\r\nP indicates a presidential election year.\r\nThe first statewide record of party affiliations was reported in 1922.\r\nHISTORICAL VOTER REGISTRATION AND\r\nPARTICIPATION IN STATEWIDE GENERAL ELECTIONS 1910-2016 (continued)\r\nS indicates a statewide special election.\r\n"
This can be useful for non-tabular content, getting a sense of the document's contents, or troubleshooting the main extraction function (e.g., sometimes there is non-visible text that confuses extract_tables()). extract_metadata() returns a list of the PDF's embedded document metadata:
str(extract_metadata(tmp))## List of 10
## $ pages : int 2
## $ title : chr "Statement of Vote - General Election, November 8, 2016"
## $ author : NULL
## $ subject : chr "Statement of Vote - General Election, November 8, 2016"
## $ keywords: chr "Statement of Vote - General Election, November 8, 2016"
## $ creator : chr "Acrobat PDFMaker 11 for Excel"
## $ producer: chr "Adobe PDF Library 11.0"
## $ created : chr "Fri Dec 16 18:54:13 GMT 2016"
## $ modified: chr "Fri Dec 16 18:54:44 GMT 2016"
## $ trapped : NULL
The make_thumbnails() function produces images (by default PNG) of pages, which can also be useful for debugging or just for the mundane purpose of image conversion:
thumb <- make_thumbnails(tmp, pages =1)library("png")
thispng <- readPNG(thumb, native =TRUE)
d <- get_page_dims(tmp, pages =1)[[1]]
plot(c(0, d[1]),c(0, d[2]), type ="n", xlab ="", ylab ="", asp =1)
rasterImage(thispng,0,0, d[1], d[2])
And, lastly, the split_pdf() and merge_pdf() functions can extract specific pages from a PDF or merge multiple PDFs together. Those functions should find multiple uses cases beyond the challenges of working with other peoples' data.
Conclusion
tabulizer can't solve all your PDF problems. More likely than not you'll at some point encounter a PDF that contains scanned tables or tables that tabula-java's algorithms can't identify well. But for a wide array of well-formatted PDF tables, tabulizer should provide a much simpler and much faster initial extraction of data than attempting to transcribe their contents manually.
Contribute
As always, the issue tracker on Github is open for suggestions, bug reports, and package support. Pull requests are always welcome.
I've flagged some specific issues on GitHub which interested users might want to help out with. These range from some basic issues:
- Identifying and creating example use cases for the new
tabulizerwiki to showcase how the package works - Adding comprehensive, cross-platform installation instructions to deal with the various intricacies of Java and rJava on various platforms
To moderately difficult issues, like:
To more advanced topics that more experienced developers - especially those with Java experience - might be interested in working on:
- Improving handling of non-latin encodings including adding tests thereof
- Preparing
tabulizerfor the migration of the tabula-java library to PDFBox 2.0, which will change some of the underlying classes (and methods thereof) thattabulizercalls from both tabula-java and PDFBox
Help of any kind on these issues will be very useful for getting the package ready for CRAN release!
Acknowledgments
Many, many thanks to the Tabula team who have done considerable work to make the tabula-java library on which tabulizer depends. I also want to express considerable thanks to David Gohel and Lincoln Mullen for their feedback during the rOpenSci onboarding process, which resulted in numerous improvements to the package and its usability, not least of which is the interactive shiny widget. Thanks, too, to Scott Chamberlain for overseeing the review process and to the whole of rOpenSci for their support of the R community.